Baby’s first Illumina MiSeq submission!
Today I submitted my first soil microbial DNA sample for Illumina sequencing! I am very excited / nervous. It’s the samples from my field trial this fall, where I had biochar additions and sorghum-sudan grass as treatments for a 2×2 experimental design, plus an additional treatment with the original, uncharred biomass, with 8 field replicates of each, at 3 timepoints. It looked something like the image I show here: a tiny tube with 0.023 mL of water that is supposed to now contain millions of microbial genetic sequences, all “barcoded” with identifying genetic sequences so I can tell the 120 unique samples apart. I am so thankful for all the support and help (both intellectually and in terms of resources) from Dan Buckley and Janice Thies‘s labs, especially from Chantal!
UPDATE: Got the fragment length quality control data back – looks pretty good. You can see some peak height variation in the target region, likely indicating varying fragment lengths across different taxa in my sample. Next step: Sequencing!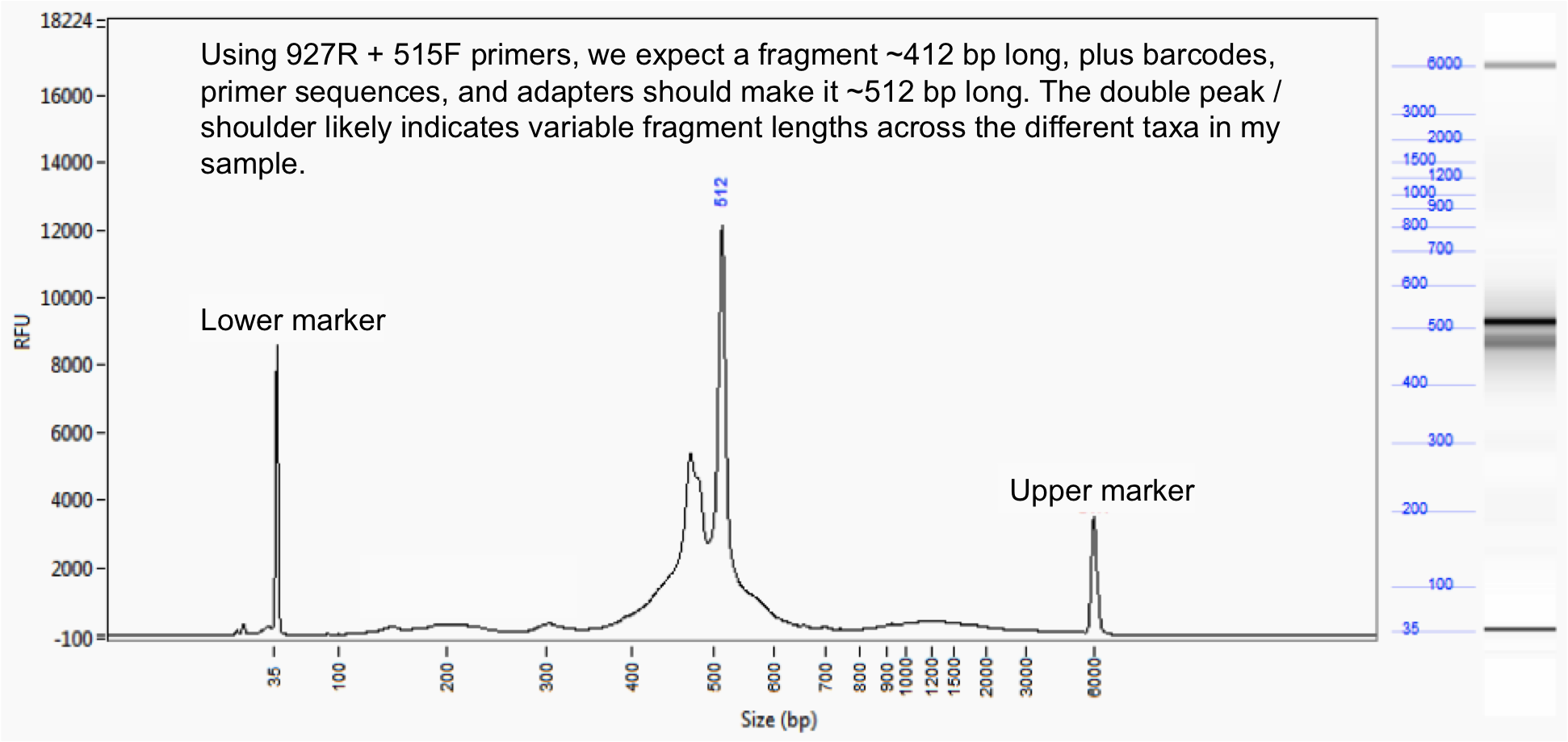
Fragment length analysis performed by Cornell Biotechnology Resource Centre
This article was posted in Uncategorized and tagged 16S, environmental community analysis, field trial, fragment length analysis, Illumina, MiSeq, sequencing, V4V5.
